|
Alomone Labs
anti ca v pan α 1 subunit antibody Anti Ca V Pan α 1 Subunit Antibody, supplied by Alomone Labs, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/anti ca v pan α 1 subunit antibody/product/Alomone Labs Average 93 stars, based on 1 article reviews
anti ca v pan α 1 subunit antibody - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
Cell Signaling Technology Inc
rabbit antimlc Rabbit Antimlc, supplied by Cell Signaling Technology Inc, used in various techniques. Bioz Stars score: 96/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/rabbit antimlc/product/Cell Signaling Technology Inc Average 96 stars, based on 1 article reviews
rabbit antimlc - by Bioz Stars,
2026-02
96/100 stars
|
Buy from Supplier |
|
Cell Signaling Technology Inc
rabbit anti mk3 antibody  Rabbit Anti Mk3 Antibody, supplied by Cell Signaling Technology Inc, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/rabbit anti mk3 antibody/product/Cell Signaling Technology Inc Average 93 stars, based on 1 article reviews
rabbit anti mk3 antibody - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
Cell Signaling Technology Inc
p stat3  P Stat3, supplied by Cell Signaling Technology Inc, used in various techniques. Bioz Stars score: 96/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/p stat3/product/Cell Signaling Technology Inc Average 96 stars, based on 1 article reviews
p stat3 - by Bioz Stars,
2026-02
96/100 stars
|
Buy from Supplier |
|
Cell Signaling Technology Inc
immunofluorescence b liu  Immunofluorescence B Liu, supplied by Cell Signaling Technology Inc, used in various techniques. Bioz Stars score: 94/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/immunofluorescence b liu/product/Cell Signaling Technology Inc Average 94 stars, based on 1 article reviews
immunofluorescence b liu - by Bioz Stars,
2026-02
94/100 stars
|
Buy from Supplier |
|
Alomone Labs
anti ryr phospho serine 2814 badrilla  Anti Ryr Phospho Serine 2814 Badrilla, supplied by Alomone Labs, used in various techniques. Bioz Stars score: 90/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/anti ryr phospho serine 2814 badrilla/product/Alomone Labs Average 90 stars, based on 1 article reviews
anti ryr phospho serine 2814 badrilla - by Bioz Stars,
2026-02
90/100 stars
|
Buy from Supplier |
|
Alomone Labs
rabbit anti kcnq1 antibody ![(a) Cartoon showing Q1/E1-YFP complex co-expressed with or without free PKA Cα subunit. (b) Representative immunoblots of lysates from HEK293 cells co-expressing Q1/E1-YFP with either empty pcDNA3.1 vector or free Cα. Anti-pKCNQ1 (top) detects phosphorylated <t>KCNQ1-S27,</t> anti-KCNQ1 (middle) detects total KCNQ1, and anti-Actin (bottom) detects total actin. (c) I Ks activation curves in CHO cells co-expressing Q1, E1-YFP with either empty pcDNA3.1 vector (black symbols, n = 13) or free PKA Cα (red symbols, n = 13). (d) Tail-decay times for currents recorded from cells co-expressing Q1/E-YFP + yotiao and either nano or free PKA Cα, or cells co-expressing Q1[S27A]/E1-YFP + yotiao and free PKA Cα. (e-h) Cartoon, immunoblots, I Ks activation curves, and population current densities of Q1/E1-YFP complex expressed with either nano (n = 10) or nanoCα ( n = 10). (i) Cartoon showing targeting of nanoCα to Q1/E1 complex via YFP tag on Q1 C-terminus. (j) Exemplar current traces I Ks traces from CHO cells co-expressing Q1-YFP/E1 with either nanoCα ( left ) or catalytically inactive nanoCα [T198A] mutant ( right ). (k) Population current densities (nano, n = 26; nanoCα, n = 19; nanoCα[T198A], n = 10).](https://bio-rxiv-images-cdn.bioz.com/dois_ending_with_26/10__1101_slash_2022__09__12__507626/10__1101_slash_2022__09__12__507626___F2.large.jpg) Rabbit Anti Kcnq1 Antibody, supplied by Alomone Labs, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/rabbit anti kcnq1 antibody/product/Alomone Labs Average 93 stars, based on 1 article reviews
rabbit anti kcnq1 antibody - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
Abcam
rabbit monoclonal anti erk1 ![(a) Cartoon showing Q1/E1-YFP complex co-expressed with or without free PKA Cα subunit. (b) Representative immunoblots of lysates from HEK293 cells co-expressing Q1/E1-YFP with either empty pcDNA3.1 vector or free Cα. Anti-pKCNQ1 (top) detects phosphorylated <t>KCNQ1-S27,</t> anti-KCNQ1 (middle) detects total KCNQ1, and anti-Actin (bottom) detects total actin. (c) I Ks activation curves in CHO cells co-expressing Q1, E1-YFP with either empty pcDNA3.1 vector (black symbols, n = 13) or free PKA Cα (red symbols, n = 13). (d) Tail-decay times for currents recorded from cells co-expressing Q1/E-YFP + yotiao and either nano or free PKA Cα, or cells co-expressing Q1[S27A]/E1-YFP + yotiao and free PKA Cα. (e-h) Cartoon, immunoblots, I Ks activation curves, and population current densities of Q1/E1-YFP complex expressed with either nano (n = 10) or nanoCα ( n = 10). (i) Cartoon showing targeting of nanoCα to Q1/E1 complex via YFP tag on Q1 C-terminus. (j) Exemplar current traces I Ks traces from CHO cells co-expressing Q1-YFP/E1 with either nanoCα ( left ) or catalytically inactive nanoCα [T198A] mutant ( right ). (k) Population current densities (nano, n = 26; nanoCα, n = 19; nanoCα[T198A], n = 10).](https://bio-rxiv-images-cdn.bioz.com/dois_ending_with_26/10__1101_slash_2022__09__12__507626/10__1101_slash_2022__09__12__507626___F2.large.jpg) Rabbit Monoclonal Anti Erk1, supplied by Abcam, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/rabbit monoclonal anti erk1/product/Abcam Average 93 stars, based on 1 article reviews
rabbit monoclonal anti erk1 - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
Abcam
ab28842 ![(a) Cartoon showing Q1/E1-YFP complex co-expressed with or without free PKA Cα subunit. (b) Representative immunoblots of lysates from HEK293 cells co-expressing Q1/E1-YFP with either empty pcDNA3.1 vector or free Cα. Anti-pKCNQ1 (top) detects phosphorylated <t>KCNQ1-S27,</t> anti-KCNQ1 (middle) detects total KCNQ1, and anti-Actin (bottom) detects total actin. (c) I Ks activation curves in CHO cells co-expressing Q1, E1-YFP with either empty pcDNA3.1 vector (black symbols, n = 13) or free PKA Cα (red symbols, n = 13). (d) Tail-decay times for currents recorded from cells co-expressing Q1/E-YFP + yotiao and either nano or free PKA Cα, or cells co-expressing Q1[S27A]/E1-YFP + yotiao and free PKA Cα. (e-h) Cartoon, immunoblots, I Ks activation curves, and population current densities of Q1/E1-YFP complex expressed with either nano (n = 10) or nanoCα ( n = 10). (i) Cartoon showing targeting of nanoCα to Q1/E1 complex via YFP tag on Q1 C-terminus. (j) Exemplar current traces I Ks traces from CHO cells co-expressing Q1-YFP/E1 with either nanoCα ( left ) or catalytically inactive nanoCα [T198A] mutant ( right ). (k) Population current densities (nano, n = 26; nanoCα, n = 19; nanoCα[T198A], n = 10).](https://bio-rxiv-images-cdn.bioz.com/dois_ending_with_26/10__1101_slash_2022__09__12__507626/10__1101_slash_2022__09__12__507626___F2.large.jpg) Ab28842, supplied by Abcam, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/ab28842/product/Abcam Average 93 stars, based on 1 article reviews
ab28842 - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
Abcam
rad21  Rad21, supplied by Abcam, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/rad21/product/Abcam Average 93 stars, based on 1 article reviews
rad21 - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
Abcam
anti alpha synuclein  Anti Alpha Synuclein, supplied by Abcam, used in various techniques. Bioz Stars score: 95/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/anti alpha synuclein/product/Abcam Average 95 stars, based on 1 article reviews
anti alpha synuclein - by Bioz Stars,
2026-02
95/100 stars
|
Buy from Supplier |
|
Abcam
phospho egfr  Phospho Egfr, supplied by Abcam, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/phospho egfr/product/Abcam Average 93 stars, based on 1 article reviews
phospho egfr - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
Image Search Results

Journal: Life Science Alliance
Article Title: FAT10 is phosphorylated by IKKβ to inhibit the antiviral type-I interferon response
doi: 10.26508/lsa.202101282
Figure Lengend Snippet: (A) The expression of endogenous FAT10 was stimulated by treating HEK293 cells with TNF/IFNγ for 24 h, followed by immunoprecipitation (IP) with a monoclonal anti-FAT10 antibody (4F1), SDS–PAGE and Coomassie blue staining (left panel). Endogenous FAT10 was cut out and sent for a phospho-proteomic analysis. As a control, samples were additionally analyzed by immunoblotting (IB) (right panel). Endogenous FAT10 was visualized with a FAT10-reactive, rabbit polyclonal antibody . β-Actin was used as a loading control. Asterisks mark the heavy and light chains of the FAT10-reactive antibody used for the immunoprecipitation. (B) Ribbon diagram of FAT10 showing the phosphorylated amino acids Ser62, Ser64, Thr77, Ser95, and Ser109 in the N- (red) and C- (blue) ubiquitin-like domain, respectively. (C) Radiolabeled phosphate was incorporated into recombinant FAT10 or ubiquitin during incubation with the recombinant kinases PINK1 or MAPKAPK3 (MK3) at 30°C for 15 or 45 min. The autoradiogram shows substrate specificity of MAPKAPK3 and PINK1 to phosphorylate FAT10 and ubiquitin, respectively. One representative experiment out of three independent experiments with similar outcomes is shown. Source data are available for this figure.
Article Snippet: Antibodies used for immunoblotting are as follows: mouse anti-FLAG antibody (F1804, 1:3,000; Merck), mouse anti-FLAG (HRP) antibody (A8592, 1:3,000; Merck), rabbit anti-FLAG antibody (F7425, 1:750; Merck), mouse anti-HA antibody (H3663, 1:5,000; Merck), rabbit anti-HA antibody (H608, 1:1,000; Merck), rabbit anti-GAPDH antibody (G9545, 1:10,000; Merck), mouse anti-tubulin antibody (T6557, 1:10,000; Merck), rabbit anti-FAT10 antibody ([ ] 1:750),
Techniques: Expressing, Immunoprecipitation, SDS Page, Staining, Control, Western Blot, Ubiquitin Proteomics, Recombinant, Incubation

Journal: Life Science Alliance
Article Title: FAT10 is phosphorylated by IKKβ to inhibit the antiviral type-I interferon response
doi: 10.26508/lsa.202101282
Figure Lengend Snippet: (A) His-3xFLAG-FAT10 (FLAG-FAT10), HA-tagged MK3, HA-MK3 TT/EE (constitutively active mutant), or HA-MK3 TT/AA (inactive mutant), were transiently overexpressed in HEK293 cells for 24 h, followed by lysis and immunoprecipitation (IP) with a monoclonal phosphoserine-reactive antibody. Subsequently, an immunoblot (IB) was performed using the antibodies indicated. Where indicated, cells were stimulated with TNF/IFNγ for 24 h before harvesting and lysis. Moreover, cells were starved for 24 h (0.3% FCS DMEM) followed by TPA treatment (30 min) before lysis, where indicated. Calf intestinal alkaline phosphatase phosphatase was added to the lysates used in lanes 3 and 5, 2 h before performing the immunoprecipitation. (B) HEK293 cells were transiently transfected with an expression plasmid for FLAG-tagged FAT10 and where indicated, additionally treated with TNF/IFNγ for 24 h. Endogenous FAT10 expression was induced by treating HEK293 cells with TNF/IFNγ for 24 h. Where indicated, lysates were incubated with 400 U of λ phosphatase for 30 min at 30°C, before the immunoprecipitation was performed. Subsequently, an immunoprecipitation against FAT10 was performed using a monoclonal FAT10-reactive antibody (clone 4F1, ) coupled to protein A sepharose, followed by Phos-tag/SDS–PAGE and IB analysis with the antibodies indicated. γ-tubulin was used as loading control. (C) Cells were prepared as in (A) and treated as specified, followed by FLAG-IP, Phos-tag/SDS–PAGE, and IB analysis with the indicated antibodies. One representative example out of three independent experiments with similar outcomes is shown. Source data are available for this figure.
Article Snippet: Antibodies used for immunoblotting are as follows: mouse anti-FLAG antibody (F1804, 1:3,000; Merck), mouse anti-FLAG (HRP) antibody (A8592, 1:3,000; Merck), rabbit anti-FLAG antibody (F7425, 1:750; Merck), mouse anti-HA antibody (H3663, 1:5,000; Merck), rabbit anti-HA antibody (H608, 1:1,000; Merck), rabbit anti-GAPDH antibody (G9545, 1:10,000; Merck), mouse anti-tubulin antibody (T6557, 1:10,000; Merck), rabbit anti-FAT10 antibody ([ ] 1:750),
Techniques: Mutagenesis, Lysis, Immunoprecipitation, Western Blot, Transfection, Expressing, Plasmid Preparation, Incubation, SDS Page, Control

Journal: Life Science Alliance
Article Title: FAT10 is phosphorylated by IKKβ to inhibit the antiviral type-I interferon response
doi: 10.26508/lsa.202101282
Figure Lengend Snippet: (A) HEK293 cells were transiently transfected with a His-3xFLAG-FAT10 (FLAG-FAT10) expression construct and stimulated for 24 h with TNF. Lysates were subjected to immunoprecipitation using FLAG-reactive antibodies, coupled to sepharose beads, and subsequently analyzed by Phos-tag/SDS–PAGE/IB analysis. Where indicated, cells were pretreated before TNF stimulation with the displayed inhibitors for a total of 3 h (10 µM each). (B) HEK293 cells were transiently transfected with expression plasmids for the different kinases. Cells were harvested, lysed, and subjected to immunoprecipitation using anti-FLAG or anti-HA antibodies, coupled to sepharose beads. Subsequently, the immunoprecipitated kinases were incubated with recombinant FAT10 (rFAT10) and an in vitro reaction was performed in the kinase buffer. The phosphorylation status of FAT10 was analyzed by Phos-tag/SDS–PAGE and IB. Asterisks mark unspecific background bands. (C) FLAG-FAT10 and the indicated kinases were transiently overexpressed in HEK293 cells followed by TNF stimulation. After 24 h, cells were lysed and subjected to immunoprecipitation against the FLAG-tag, combined with Phos-tag/SDS–PAGE and IB analysis. (D) Recombinant FAT10 (rFAT10) was incubated with recombinant kinases IKKβ, IKKε or MK3 for 45 min at 30°C. Subsequently, proteins were separated on a Phos-tag/SDS–PAGE followed by immunoblot analysis using the antibodies indicated. (E) HEK293 cells were prepared as described in (A). Where specified, cells were pretreated with the inhibitors indicated (10 µM each) for a total of 3 h before stimulation with TNF. One representative example out of three independent experiments with the same outcomes is shown. Source data are available for this figure.
Article Snippet: Antibodies used for immunoblotting are as follows: mouse anti-FLAG antibody (F1804, 1:3,000; Merck), mouse anti-FLAG (HRP) antibody (A8592, 1:3,000; Merck), rabbit anti-FLAG antibody (F7425, 1:750; Merck), mouse anti-HA antibody (H3663, 1:5,000; Merck), rabbit anti-HA antibody (H608, 1:1,000; Merck), rabbit anti-GAPDH antibody (G9545, 1:10,000; Merck), mouse anti-tubulin antibody (T6557, 1:10,000; Merck), rabbit anti-FAT10 antibody ([ ] 1:750),
Techniques: Transfection, Expressing, Construct, Immunoprecipitation, SDS Page, Incubation, Recombinant, In Vitro, Phospho-proteomics, FLAG-tag, Western Blot

Journal: International Journal of Molecular Medicine
Article Title: GFI1 promotes the proliferation and migration of esophageal squamous cell carcinoma cells through the inhibition of SOCS1 expression
doi: 10.3892/ijmm.2021.5017
Figure Lengend Snippet: GFI1 enhances NF-κB and STAT3 activity by inhibiting SOCS1 expression in esophageal squamous cell carcinoma cell lines. (A and B) siGFI1 was transfected into KYSE30 and KYSE150 cells. (A) mRNA and (B) protein expression levels of SOCS1 were examined using reverse transcription-quantitative PCR and western blotting, respectively. (C) NF-κB activity, (D) p-p65 and (E) p-STAT3 protein levels were analyzed in KYSE30/KYSE150 cells following transfection with siGFI1. * P<0.05, ** P<0.01 and *** P<0.001 vs. control. si, small interfering RNA; GFI1, growth factor-independent 1; SOCS1, suppressor of cytokine signaling 1; p-, phosphorylated.
Article Snippet: Rabbit polyclonal antibodies against phosphorylated (p)-p65 (1:1,000; cat. no. 310012) were obtained from Chengdu Zen Bioscience Co., Ltd. Rabbit monoclonal antibodies against STAT3 (1:1,000; cat. no. 4904S) and
Techniques: Activity Assay, Expressing, Transfection, Reverse Transcription, Real-time Polymerase Chain Reaction, Western Blot, Control, Small Interfering RNA

Journal: International Journal of Molecular Medicine
Article Title: GFI1 promotes the proliferation and migration of esophageal squamous cell carcinoma cells through the inhibition of SOCS1 expression
doi: 10.3892/ijmm.2021.5017
Figure Lengend Snippet: SOCS1 inhibits the increase in esophageal squamous cell carcinoma cell proliferation and migration induced by GFI1. (A-H) siSOCS1 was transfected into the GFI1-knockdown KYSE30 and KYSE150 cells. (A) Colony formation assays were performed on the double transfected cells. (B) Cell migration was determined using Transwell assays. (C) NF-κB activity, (D) p-p65, p-STAT3 and SOCS1 protein levels were analyzed. (E-G) Ratio of p-p65 to p65 and STAT3 to p-STAT3, as well as (H) the semi-quantification of SOCS1 protein expression were determined using ImageJ. Magnification, ×200. * P<0.05, ** P<0.01 *** P<0.001 vs. control; # P<0.05, ## P<0.01 and ### P<0.001 vs. siGFI1. SOCS1, suppressor of cytokine signaling 1; si, small interfering RNA; GFI1, growth factor-independent 1; p-, phosphorylated.
Article Snippet: Rabbit polyclonal antibodies against phosphorylated (p)-p65 (1:1,000; cat. no. 310012) were obtained from Chengdu Zen Bioscience Co., Ltd. Rabbit monoclonal antibodies against STAT3 (1:1,000; cat. no. 4904S) and
Techniques: Migration, Transfection, Knockdown, Activity Assay, Expressing, Control, Small Interfering RNA

Journal: International Journal of Molecular Medicine
Article Title: GFI1 promotes the proliferation and migration of esophageal squamous cell carcinoma cells through the inhibition of SOCS1 expression
doi: 10.3892/ijmm.2021.5017
Figure Lengend Snippet: Regulatory model of the role of GFI1 in ESCC progression. The results of the present study demonstrated that the upregulation of GFI1 promoted ESCC proliferation and migration via inhibition of SOCS1 expression, leading to the activation of the NF-κB and STAT3 pathways. GFI1, growth factor-independent 1; ESCC, esophageal squamous cell carcinoma; SOCS1, suppressor of cytokine signaling 1.
Article Snippet: Rabbit polyclonal antibodies against phosphorylated (p)-p65 (1:1,000; cat. no. 310012) were obtained from Chengdu Zen Bioscience Co., Ltd. Rabbit monoclonal antibodies against STAT3 (1:1,000; cat. no. 4904S) and
Techniques: Migration, Inhibition, Expressing, Activation Assay
![(a) Cartoon showing Q1/E1-YFP complex co-expressed with or without free PKA Cα subunit. (b) Representative immunoblots of lysates from HEK293 cells co-expressing Q1/E1-YFP with either empty pcDNA3.1 vector or free Cα. Anti-pKCNQ1 (top) detects phosphorylated KCNQ1-S27, anti-KCNQ1 (middle) detects total KCNQ1, and anti-Actin (bottom) detects total actin. (c) I Ks activation curves in CHO cells co-expressing Q1, E1-YFP with either empty pcDNA3.1 vector (black symbols, n = 13) or free PKA Cα (red symbols, n = 13). (d) Tail-decay times for currents recorded from cells co-expressing Q1/E-YFP + yotiao and either nano or free PKA Cα, or cells co-expressing Q1[S27A]/E1-YFP + yotiao and free PKA Cα. (e-h) Cartoon, immunoblots, I Ks activation curves, and population current densities of Q1/E1-YFP complex expressed with either nano (n = 10) or nanoCα ( n = 10). (i) Cartoon showing targeting of nanoCα to Q1/E1 complex via YFP tag on Q1 C-terminus. (j) Exemplar current traces I Ks traces from CHO cells co-expressing Q1-YFP/E1 with either nanoCα ( left ) or catalytically inactive nanoCα [T198A] mutant ( right ). (k) Population current densities (nano, n = 26; nanoCα, n = 19; nanoCα[T198A], n = 10).](https://bio-rxiv-images-cdn.bioz.com/dois_ending_with_26/10__1101_slash_2022__09__12__507626/10__1101_slash_2022__09__12__507626___F2.large.jpg)
Journal: bioRxiv
Article Title: Divergent regulation of KCNQ1/E1 by targeted recruitment of protein kinase A to distinct sites on the channel complex
doi: 10.1101/2022.09.12.507626
Figure Lengend Snippet: (a) Cartoon showing Q1/E1-YFP complex co-expressed with or without free PKA Cα subunit. (b) Representative immunoblots of lysates from HEK293 cells co-expressing Q1/E1-YFP with either empty pcDNA3.1 vector or free Cα. Anti-pKCNQ1 (top) detects phosphorylated KCNQ1-S27, anti-KCNQ1 (middle) detects total KCNQ1, and anti-Actin (bottom) detects total actin. (c) I Ks activation curves in CHO cells co-expressing Q1, E1-YFP with either empty pcDNA3.1 vector (black symbols, n = 13) or free PKA Cα (red symbols, n = 13). (d) Tail-decay times for currents recorded from cells co-expressing Q1/E-YFP + yotiao and either nano or free PKA Cα, or cells co-expressing Q1[S27A]/E1-YFP + yotiao and free PKA Cα. (e-h) Cartoon, immunoblots, I Ks activation curves, and population current densities of Q1/E1-YFP complex expressed with either nano (n = 10) or nanoCα ( n = 10). (i) Cartoon showing targeting of nanoCα to Q1/E1 complex via YFP tag on Q1 C-terminus. (j) Exemplar current traces I Ks traces from CHO cells co-expressing Q1-YFP/E1 with either nanoCα ( left ) or catalytically inactive nanoCα [T198A] mutant ( right ). (k) Population current densities (nano, n = 26; nanoCα, n = 19; nanoCα[T198A], n = 10).
Article Snippet: Membranes were pre-treated with 0.5% glutaraldehyde and re-blotted with
Techniques: Western Blot, Expressing, Plasmid Preparation, Activation Assay, Mutagenesis
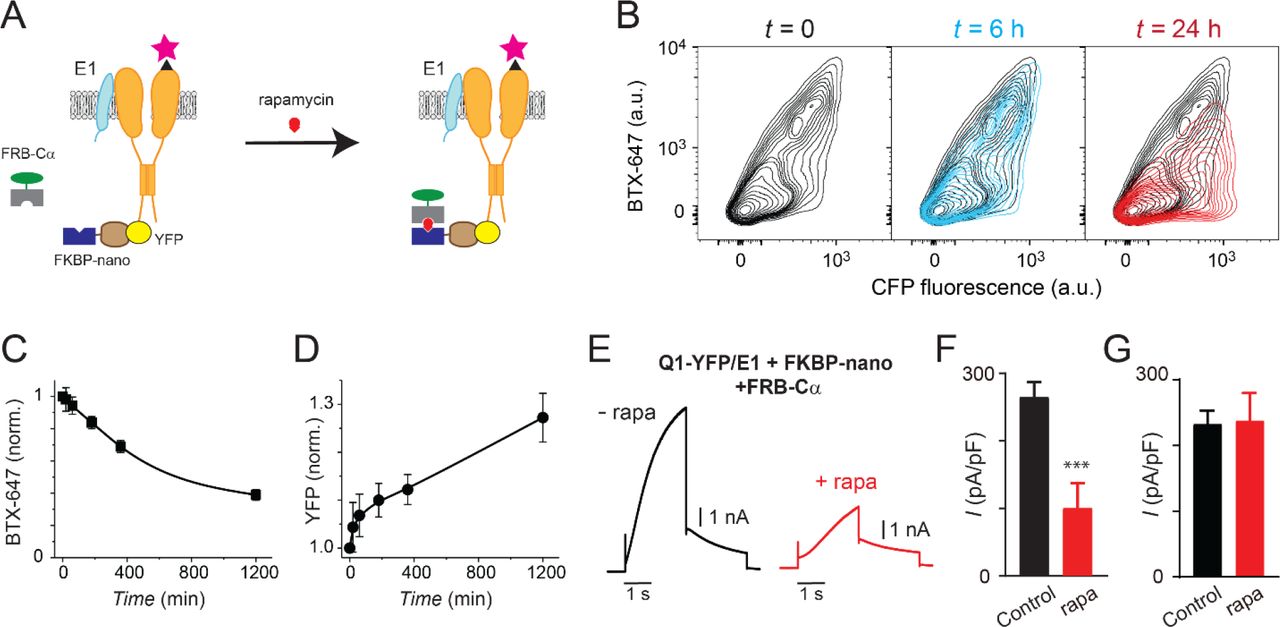
Journal: bioRxiv
Article Title: Divergent regulation of KCNQ1/E1 by targeted recruitment of protein kinase A to distinct sites on the channel complex
doi: 10.1101/2022.09.12.507626
Figure Lengend Snippet: (a) Cartoon of FKBP/FRB heterodimerization strategy utilized for rapamycin-induced recruitment of engineered Cα to BBS-Q1-YFP/E1. (B) Exemplar flow cytometry contour plots showing surface expression (BTX-647 fluorescence) and CFP fluorescence in cells expressing BBS-Q1-YFP/E1 with FRB-Cα and FKBP-nano at times t = 0 ( left ), t = 6h ( middle ) and t = 24h ( right ) after rapamycin addition. (c) Normalized mean Q1 surface density (BTX-647 fluorescence) plotted as a function of time after rapamycin induction. (d) Normalized mean Q1 total expression (YFP fluorescence) plotted as a function of time after rapamycin induction. (e) Exemplar I Ks traces recorded in CHO cells co-expressing KCNQ1-YFP/KCNE1/Nano-FKBP-FRB-Cα incubated 20 hours either without ( left ) or with ( right ) rapamycin. (f) Mean current densities in CHO cells co-expressing KCNQ1-YFP/KCNE1/Nano-FKBP-FRB-Cα without rapamycin (black, n = 10) or after 20 h rapamycin incubation (red, n = 14). *** P < 0.001, paired t test. (g) Mean current densities in control cells co-expressing KCNQ1-YFP/KCNE1 without rapamycin (black, n = 8) or after 20 h rapamycin incubation (red, n = 9).
Article Snippet: Membranes were pre-treated with 0.5% glutaraldehyde and re-blotted with
Techniques: Flow Cytometry, Expressing, Fluorescence, Incubation, Control
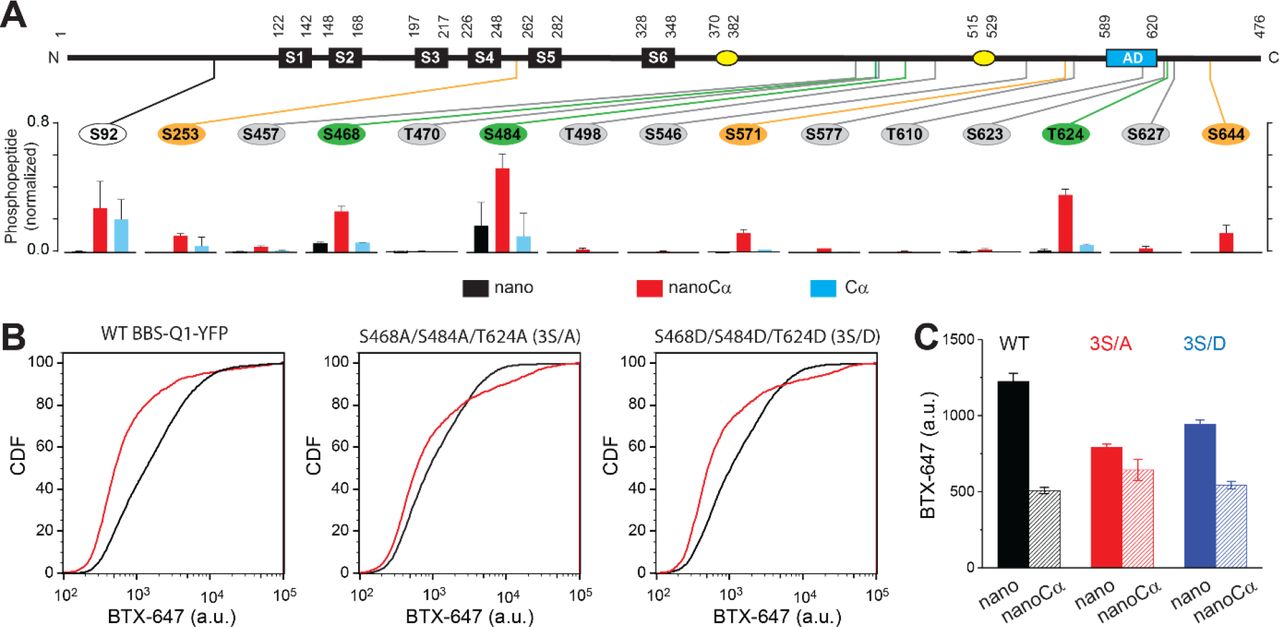
Journal: bioRxiv
Article Title: Divergent regulation of KCNQ1/E1 by targeted recruitment of protein kinase A to distinct sites on the channel complex
doi: 10.1101/2022.09.12.507626
Figure Lengend Snippet: (a) Top , schematic of Q1 showing positions of Ser and Thr residues where phosphorylation was increased when nanoCα was targeted to Q1 C-terminus. Bottom , relative abundance of phosphorylated KCNQ1-YFP peptides identified using mass spectrometry in cells co-expressing nano (black), nanoCα (red), or free Cα (cyan). (b) Exemplar CDF plots showing channel surface density in cells expressing WT BBS-Q1-YFP ( left ), BBS-3S/A-YFP ( middle ), or BBS-3S/D-YFP ( right ) in the absence (black traces) or presence (red traces) of nanoCα. (c) Channel surface density (BTX-647 fluorescence) in cells expressing WT BBS-Q1-YFP, BBS-3S/A-YFP, or BBS-3S/D-YFP in the presence of either nano or nanoCα. WT BBS-Q1-YFP (nano, n = 15,411, N = 2; nanoCα, n = 9,053, N = 2). BBS-3S/A-YFP (nano, n = 6,158, N = 2; nanoCα, n = 4,798, N = 2). BBS-3S/D-YFP (nano, n = 11,823, N = 2; nanoCα, n = 5,685, N = 2).
Article Snippet: Membranes were pre-treated with 0.5% glutaraldehyde and re-blotted with
Techniques: Phospho-proteomics, Mass Spectrometry, Expressing, Fluorescence

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) E17.5-E18.5 cortices were dissociated and plated on poly-D-lysine. After 10d, cultures were stained for pan neuronal (MAP2), astrocyte (GFAP) and microglia (IBA1) markers, and cell type composition was determined by quantitative analysis of immunofluorescence images. Based on 6 Rad21 +/+ Nex Cre and 8 Rad21 lox/lox Nex Cre different samples analysed in 4 independent experiments. b) Immunofluorescence staining of Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre neuronal explant cultures for RAD21 and MAP2 (left) and distribution of RAD21 expression by MAP + neurons (right). Note the discontinuous distribution of RAD21 expression in Rad21 lox/lox Nex Cre neurons. Three independent experiments per genotype. DAPI marks nuclei. Scale bar = 60 μm. c) Immunofluorescence staining for RAD21, MAP2, and the marker of GABAergic inhibitory neurons, GAD67 (left). Distribution of RAD21 expression in GAD67 + and GAD67 - neurons (right). Note that the discontinuous distribution of RAD21 expression in Rad21 lox/lox Nex Cre neuronal explant cultures is due to GAD67 + GABAergic inhibitory neurons. Three independent experiments for Rad21 +/+ Nex Cre and 6 independent experiments for Rad21 lox/lox Nex Cre . DAPI marks nuclei. Scale bar = 20 μm. d) Quantitative RT-PCR analysis of Rad21 mRNA expression in Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre cortical explant cultures (mean ± SEM, n=18). Hprt and Ubc were used for normalization (left). RAD21 protein expression in Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre cortical explant cultures was quantified by fluorescent immunoblots (mean ± SEM, n=6) and normalised to LaminB (center). Nex Cre RiboTag RNA-seq of analysis of Rad21 mRNA expression in Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre cortical explant cultures (right, 3 independent biological replicates). e) 5C heat maps of a 1.72 Mb region on chromosome 2, comparing Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre cortical explant cultures. CTCF ChIP-seq (Ref. ) and mm9 coordinates are shown for reference. Arrowheads mark the position of CTCF-based loops. Results were consistent across two replicates and 3 chromosomal regions Histograms below show the quantification of representative CTCF-based loops (arrowheads) in two independent biological replicates for control and Rad21 lox/lox Nex Cre neurons.
Article Snippet: Primary antibodies used were specific to
Techniques: Staining, Immunofluorescence, Expressing, Marker, Quantitative RT-PCR, Western Blot, RNA Sequencing Assay, ChIP-sequencing

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Nex Cre -dependent Rpl22-HA (RiboTag) expression is restricted to RAD21-negative cells in Rad21 lox/lox Nex Cre neurons. Immunofluorescence staining for RAD21, the pan-neuronal marker MAP2 and HA (RiboTag) in explant culture. DAPI marks nuclei. Scale bar = 40 μm. b) Nex Cre RiboTag captures excitatory neuron-specific transcripts such as Slc17a7 and Camk2a and depletes cell type-specific transcripts expressed in inhibitory neurons ( Gad1, Gad2, Slc32a1 ), astrocytes ( Gfap, Aqp4, Mlc1 ), and microglia ( Aif ). Transcript enrichment (or depletion) was calculated using the normalized counts from Nex Cre RiboTag versus standard RNA-seq in Rad21 +/+ Nex Cre neurons.
Article Snippet: Primary antibodies used were specific to
Techniques: Expressing, Immunofluorescence, Staining, Marker, RNA Sequencing Assay

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Volcano plot representing log2 fold-change (FC) versus significance (-log10 of adjusted P values) of downregulated genes (1028) and upregulated genes (572) in RiboTag RNA-seq of Rad21 lox/lox Nex Cre versus Rad21 +/+ Nex Cre neurons. Red marks Rad21 . b) Analysis of gene ontology of biological functions of deregulated genes in Rad21 lox/lox Nex Cre neurons. Enrichment is calculated relative to expressed genes. c) The percentage of constitutive and activity-dependent genes deregulated in Rad21 lox/lox Nex Cre neurons in explant culture at baseline as determined by RiboTag RNA-seq. The P -value (Fisher Exact Test) and Odds ratio indicate that activity-dependent genes are more frequently deregulated than constitutive genes.
Article Snippet: Primary antibodies used were specific to
Techniques: RNA Sequencing Assay, Activity Assay

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Examples of deregulated genes in Rad21 lox/lox Nex Cre neurons. Genes associated with autism spectrum disorders are highlighted in red. b) GSEA for downregulated genes in Nex Cre/+ Rad21 lox/lox neurons using gene sets derived from (i) KEGG pathway database, (ii) GO Biological Process Ontology, (iii) GO Molecular Function Ontology, (iv) GO Cellular Component Ontology in the Molecular Signatures Database (MSigDB). c) Overlap between human genes associated with autism spectrum disorders from the SFARI database and differentially expressed genes (left), downregulated genes (middle) and upregulated genes (right) in Rad21 lox/lox Nex Cre cortical neurons.
Article Snippet: Primary antibodies used were specific to
Techniques: Derivative Assay

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Expected Mendelian ratios and observed percentages of live Rad21 +/+ Nex Cre , Rad21 lox/+ Nex Cre , Rad21 lox/lox Nex Cre mice at the indicated developmental stages, n = 217. b) Immunofluorescence analysis shows neither the apoptosis marker activated caspase 3 (CC3) nor the DNA damage marker γH2AX in E16.5 (top) and E18.5 Rad21 lox/lox Nex Cre (bottom, white lines demarcate the cortex). Wild type E16.5 thymi are shown as positive controls for CC3 and γH2AX. Two biological replicates. Scale bar = 100 μm. Photomicrographs of coronal brain sections at gestational age E16 modified from the Atlas of the prenatal mouse brain are shown for orientation. c) Quantitative RT-PCR analysis of gene expression in Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre E17.5/18.5 cortical explant cultures 10 d after plating. Hprt and Ubc were used for normalization. Mean ± SEM of 3 cultures per genotype. d) Brain weights of Rad21 +/+ Nex Cre and Rad21 lox/+ Nex Cre , Rad21 lox/lox Nex Cre mice at birth (P0). Mean ± SEM of between 3 and 13 mice per genotype. e) Embryonic cortices from wild-type and Rad21 lox/lox Nex Cre mice were dissected at E17.5 and E18.5 and dissociated. Cortical cell numbers were determined by counting in Neubauer chambers. Each symbol denotes an independent experiment. Mean ± SEM are also shown.
Article Snippet: Primary antibodies used were specific to
Techniques: Immunofluorescence, Marker, Modification, Quantitative RT-PCR, Expressing

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Schema of cortical layers showing subplate (SP), layer 6 (VI), layer 5 (V), the cortical plate (CP), and the marginal zone (MZ). Immunofluorescence analysis of the neuronal transcription factors CUX1, TBR1, and CTIP2 at E16.5. Representative of 3 biological replicates. Scale bar = 100 μm. b) Morphology of E18.5 neurons after 1 d in explant culture. Immunofluorescence staining for the pan-neuronal marker MAP2, tubulin beta 3 (TUBB3), and DAPI. Scale bar = 20 μm. c) Morphology of Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre cortical neurons in explant culture on rat glia . Cultures were sparsely labeled with GFP to visualize individual cells and their processes, and stained for GAD67 to exclude GABAergic neurons. Dendritic traces of GFP + neurons. Scale bar = 50 μm. d) Sholl analysis of Rad21 +/+ Nex Cre and Rad21 lox/lox Nex Cre cortical neurons in explant cultures shown in c). Shown is the number of crossings, dendritic length, terminal points, branch points and spines per 10 μm. Three independent experiments, 32 Rad21 lox/lox Nex Cre and 28 Rad21 +/+ Nex Cre neurons except for the number of spines (two independent experiments, 10 Rad21 lox/lox Nex Cre and 10 Rad21 +/+ Nex Cre neurons). * adj. P <0.05, ** adj. P <0.01, *** adj. P <0.001, **** adj. P <0.0001. Scale bar = 10 μm.
Article Snippet: Primary antibodies used were specific to
Techniques: Immunofluorescence, Staining, Marker, Labeling

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) GSEA of the gene set downregulated (DEseq2, adj. P < 0.05) in RAD21-TEV neurons in Rad21 lox/lox Nex Cre neurons (left) and GSEA of genes downregulated in Rad21 lox/lox Nex Cre neurons (DEseq2, adj. P < 0.05) in RAD21-TEV neurons. b) Scatter plots of gene expression within aggregate GO terms, comparing RAD21-TEV with Rad21 lox/lox Nex Cre neurons. Genes that were found deregulated in at least one of the genotypes are shown. P -values and odds ratios refer to the probability of finding the observed patterns of co-regulation by chance. R S : Spearman’s rank coefficient. c) Deregulation of constitutive and activity-dependent genes 24h after acute cohesin depletion by inducible proteolytic cleavage of RAD21-TEV; adj. P <0.05 based on DEseq2 analysis of 3 RNA-seq replicates per experiment. Two independent experiments are shown.
Article Snippet: Primary antibodies used were specific to
Techniques: Expressing, Activity Assay, RNA Sequencing Assay

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Preferential deregulation in Rad21 lox/lox Nex Cre neurons of genes near constitutive and KCl-inducible neuronal enhancers . Based on 3 RiboTag RNA-seq replicates per genotype. b) Enrichment of inducible activity-dependent genes near constitutive and KCl-inducible neuronal enhancers . c) CTCF binding at neuronal genes and enhancers. All genes: all expressed genes in total RNA-seq; Activity-dependent genes: Previously defined activity-dependent genes (Kim et al., 2010) that are inducible by KCl in our experiments (KCl minus TTX adj. P < 0.05); Constitutive genes: Expressed genes that are not inducible by KCl in our experiments (KCl minus TTX adj. P > 0.05); Enhancers: Previously defined forebrain enhancers ; CTCF binding: Previously defined CTCF binding peaks within 1kb of TSS or enhancer. d) Only a minority of activity-dependent gene promoters are directly bound by CTCF, and most activity-dependent genes that lack CTCF promoter binding are nevertheless deregulated in cohesin-deficient neurons. e) Models of gene regulation by direct (left) versus domain-wide chromatin contacts (right). CD: contact domain.
Article Snippet: Primary antibodies used were specific to
Techniques: RNA Sequencing Assay, Activity Assay, Binding Assay

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Top: The percentage of constitutive and activity-dependent genes deregulated in Rad21 lox/lox Nex Cre neurons in explant culture at baseline as determined by RNA-seq. Analysis of previously defined activity-dependent genes . Middle: Fraction of constitutive and activity-dependent genes deregulated in Rad21 lox/lox Nex Cre neurons in the presence of TTX and D-AP5 (TTX). Bottom: Fraction of constitutive and activity-dependent genes deregulated in Rad21 lox/lox Nex Cre neurons after 6h stimulation with KCl. b) Expression of activity-dependent genes in explant cultures of Rad21 +/+ and Rad21 lox/lox Nex Cre neurons under baseline conditions that allow for cell-cell communication versus TTX/D-AP5 (TTX). Comparison by two-sample Kolmogorov-Smirnov test showed that Rad21 +/+ Nex Cre neurons showed stronger expression of activity-dependent genes than Rad21 lox/lox Nex Cre neurons ( P = 5.89e-11). c) Fraction of activity-dependent genes significantly induced by KCl in Rad21 lox/lox Nex Cre neurons at 1 and 6h. In wild-type neurons, 117 and 810 genes were induced ≥2-fold at 1 and 6h of KCl treatment, respectively. d) Fraction of activity-dependent genes that were significantly induced by BDNF at 30 and 120min in RAD21-TEV neurons 24h after RAD21 cleavage. In control RAD21-TEV neurons, 16 and 261 activity-dependent genes were induced ≥2-fold 30 and 120min after BDNF treatment, respectively. Most of these remained inducible 24h after RAD21-TEV cleavage. Dark orange: Strongly induced: log2 FC>1, adj. P <0.05; light orange: moderately induced (log2 FC > 0.5, adj. P <0.05); grey: weakly induced (adj. P <0.05); white: not induced (adj. P >0.05).
Article Snippet: Primary antibodies used were specific to
Techniques: Activity Assay, RNA Sequencing Assay, Expressing

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Expression of the activity-dependent Fos gene at baseline, after TTX/D-AP5 (TTX), and KCl-stimulation (left, mean log2-transformed counts from 3 biological replicates, * adj. P < 0.05). b) Enhancer transcripts in control and Rad21 lox/lox Nex Cre neurons were quantified based on normalized RNA-seq reads within 1kb of the eRNA transcription start site. An intergenic region on chr11 was used as a negative control (71.177.622-71.177.792). c) H3K27ac ChIP normalized to H3 in control and Rad21 lox/lox Nex Cre neurons at a control site, Fos enhancer 1 and Fos enhancer 2 after TTX/D-AP5 (TTX) or 1h KCl (KCl). d) Interaction score heatmaps of the 65 kb region immediately surrounding Fos obtained by 5C. Black frames highlight interactions between the Fos gene and upstream enhancers 1 and 2. Previously published CTCF-ChIP-seq is shown for orientation and H3K27ac ChIP-seq in inactive (TTX-treated) and activated neurons is shown to annotate enhancer regions . RNA-seq in TTX-treated and 1h KCl-activated control and Rad21 lox/lox Nex Cre neurons shows KCl-inducible transcription of Fos enhancers in wild-type and cohesin-deficient neurons. Two independent biological replicates are shown in . e) Quantification of 5C contacts between the Fos promoter and Fos enhancer 1 (top), the Fos promoter and Fos enhancer 2 (middle), and CTCF-marked boundaries of the sub-TAD containing Fos (bottom). Two replicates per genotype and condition. f) Model for how cohesin-mediated domain-wide contacts alter the probability of enhancer-promoter contacts, and in this way fine-tune the transcription of activity-dependent genes at baseline and in response to activation. In the absence of cohesin, many activity-dependent genes are expressed at inappropriate levels, but most remain responsive to inducing activation signals. At the Fos and Arc loci, cohesin is not required for chromatin contacts between inducible enhancers and their target promoters. See text for details.
Article Snippet: Primary antibodies used were specific to
Techniques: Expressing, Activity Assay, Transformation Assay, RNA Sequencing Assay, Negative Control, ChIP-sequencing, Activation Assay

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Expression of Arc mRNA at baseline, after TTX/D-AP5 (TTX), and KCl-stimulation (top, mean log2-transformed counts from 3 biological RNA-seq replicates, * adj. P < 0.05) and at the indicated time after BDNF stimulation (bottom, data points represent biological RT-PCR replicates). P- values refer to induction relative to 0 min. * P < 0.05, *** P <0.001. b) Interaction score heatmaps of the ∼40 kb region immediately surrounding Arc obtained by 5C for resting (TTX) and 1h KCl-activated wild-type (top) and Rad21 lox/lox Nex Cre neurons (bottom). Black frames highlight interaction between the Arc gene (y-axis) and a nearby downstream enhancer (x-axis). Previously published CTCF-ChIP-seq is shown . H3K27ac ChIP-seq in inactive (TTX-treated) and activated neurons is shown to annotate enhancer regions . Two independent biological replicates are shown in . c) Quantification of 5C data. Arc enhancer-promoter loop (top). A CTCF-based loop that braces the Arc locus (arrowhead marked with * in panel b) is quantified for comparison (bottom).
Article Snippet: Primary antibodies used were specific to
Techniques: Expressing, Transformation Assay, RNA Sequencing Assay, Reverse Transcription Polymerase Chain Reaction, ChIP-sequencing

Journal: bioRxiv
Article Title: Activity-induced gene expression and long-range enhancer-promoter contacts in cohesin-deficient neurons
doi: 10.1101/2021.02.24.432639
Figure Lengend Snippet: a) Top: Interaction frequency zoom-in heatmaps of 250 kb region surrounding the Fos gene. Dashed lines and arrow heads mark major CTCF binding sites at the boundaries of the domain that contains Fos . Note the weakening of these contacts in Rad21 lox/lox Nex Cre neurons. Bottom: Interaction score heatmaps of the 65 kb region immediately surrounding the Fos gene. Black frames highlight interactions between the Fos gene and upstream enhancers 1 and 2. H3K27ac ChIP-seq data from Bicuculline-treated (active) and TTX-treated (inactive) neurons annotate enhancer regions. Two independent biological replicates are shown. b) Top: Interaction frequency zoom-in heatmaps of ∼200 kb region surrounding the Arc gene. Dashed lines and arrow heads mark major CTCF binding sites at the boundaries of domains that contain the Arc locus. Note the weakening of these contacts in Rad21 lox/lox Nex Cre neurons. Bottom: Interaction score heatmaps of the ∼40 kb region immediately surrounding the Arc gene. Black frames highlight interaction between the Arc gene (y-axis) and a nearby downstream enhancer (x-axis). H3K27ac ChIP-seq data from Bicuculline-treated (active) and TTX-treated (inactive) neurons annotate enhancer regions. Two independent biological replicates are shown.
Article Snippet: Primary antibodies used were specific to
Techniques: Binding Assay, ChIP-sequencing

Journal: Translational Neurodegeneration
Article Title: Alpha-synuclein overexpression in the olfactory bulb initiates prodromal symptoms and pathology of Parkinson’s disease
doi: 10.1186/s40035-018-0128-6
Figure Lengend Snippet: Immunofluorescent double-labeled staining in the SN field. a - c double staining of SN field by LC3B (green) and TUNEL staining (red) in the control rats. d - f double staining of SN field by LC3B (green) and TUNEL staining (red) in the hm-α-synuclein rats. These data indicates that the apoptosis happen in some cells overexpressing the hm-α-synuclein. g - i double staining of SN field by ThS (green) and pSer129-α-synuclein staining (red) in the control rats. j - l double staining of SN field by ThS (green) and pSer129-α-synuclein staining (red) in the hm-α-synuclein rats. These data indicates that pSer129-α-synuclein signals reflected LB pathological changes. (Bar: 20um)
Article Snippet: To stain coronal free-floating sections, primary antibodies anti-rat α-syn (monoclonal raised in mouse, 1:500, Abcam) or anti-TH (raised in rabbit, 1:1,000,
Techniques: Labeling, Staining, Double Staining, TUNEL Assay